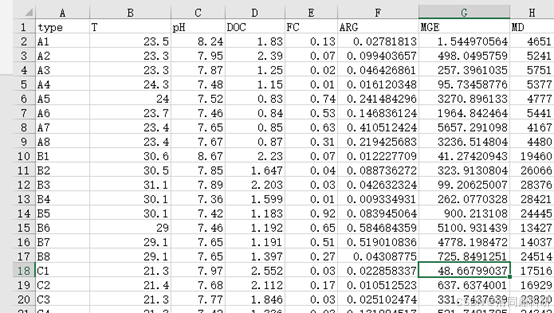
class="hljs-ln-code"> class="hljs-ln-line">setwd("E:/R/piecewiseSEM") class="hljs-ln-numbers"> class="hljs-ln-line hljs-ln-n" data-line-number="3"> class="hljs-ln-code"> class="hljs-ln-line"> class="hljs-ln-numbers"> class="hljs-ln-line hljs-ln-n" data-line-number="4"> class="hljs-ln-code"> class="hljs-ln-line"> class="hljs-ln-numbers"> class="hljs-ln-line hljs-ln-n" data-line-number="5"> class="hljs-ln-code"> class="hljs-ln-line"> class="hljs-ln-numbers"> class="hljs-ln-line hljs-ln-n" data-line-number="6"> class="hljs-ln-code"> class="hljs-ln-line">library(piecewiseSEM) class="hljs-ln-numbers"> class="hljs-ln-line hljs-ln-n" data-line-number="7"> class="hljs-ln-code"> class="hljs-ln-line"> class="hljs-ln-numbers"> class="hljs-ln-line hljs-ln-n" data-line-number="8"> class="hljs-ln-code"> class="hljs-ln-line"> class="hljs-ln-numbers"> class="hljs-ln-line hljs-ln-n" data-line-number="9"> class="hljs-ln-code"> class="hljs-ln-line"> class="hljs-ln-numbers"> class="hljs-ln-line hljs-ln-n" data-line-number="10"> class="hljs-ln-code"> class="hljs-ln-line">mydata<-read.csv("data.csv") class="hljs-ln-numbers"> class="hljs-ln-line hljs-ln-n" data-line-number="11"> class="hljs-ln-code"> class="hljs-ln-line"> class="hljs-ln-numbers"> class="hljs-ln-line hljs-ln-n" data-line-number="12"> class="hljs-ln-code"> class="hljs-ln-line"> class="hljs-ln-numbers"> class="hljs-ln-line hljs-ln-n" data-line-number="13"> class="hljs-ln-code"> class="hljs-ln-line">model <- psem( class="hljs-ln-numbers"> class="hljs-ln-line hljs-ln-n" data-line-number="14"> class="hljs-ln-code"> class="hljs-ln-line"> lm(MD~ T+FC+pH+DOC, mydata), class="hljs-ln-numbers"> class="hljs-ln-line hljs-ln-n" data-line-number="15"> class="hljs-ln-code"> class="hljs-ln-line"> lm(MGE~ MD, mydata ), class="hljs-ln-numbers"> class="hljs-ln-line hljs-ln-n" data-line-number="16"> class="hljs-ln-code"> class="hljs-ln-line"> lm(ARG~ MD, mydata ) class="hljs-ln-numbers"> class="hljs-ln-line hljs-ln-n" data-line-number="17"> class="hljs-ln-code"> class="hljs-ln-line">) class="hljs-ln-numbers"> class="hljs-ln-line hljs-ln-n" data-line-number="18"> class="hljs-ln-code"> class="hljs-ln-line"> class="hljs-ln-numbers"> class="hljs-ln-line hljs-ln-n" data-line-number="19"> class="hljs-ln-code"> class="hljs-ln-line">summary(model, .progressBar = F) class="hide-preCode-box"> class="hljs-button signin active" data-title="登录复制" data-report-click="{"spm":"1001.2101.3001.4334"}" onclick="hljs.signin(event)">
class="hljs-button signin active" data-title="登录复制" data-report-click="{"spm":"1001.2101.3001.4334"}" onclick="hljs.signin(event)">
结果如下,p=0 显然模型是不合理的。其次提示缺少一些路径:

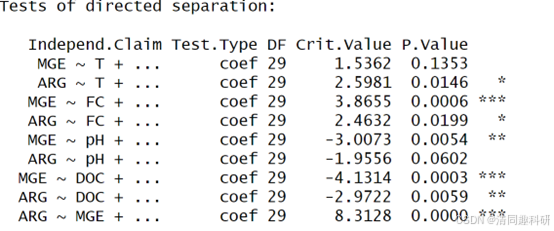
因此我们考虑,水质指标直接对抗性基因(ARG,MGE)的影响,修改模型如下:
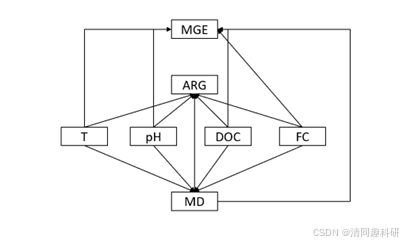
代码如下:
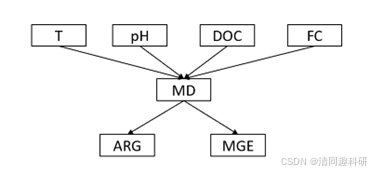
- class="hljs-ln-numbers"> class="hljs-ln-line hljs-ln-n" data-line-number="1"> class="hljs-ln-code"> class="hljs-ln-line">model <- psem(
- class="hljs-ln-numbers"> class="hljs-ln-line hljs-ln-n" data-line-number="2"> class="hljs-ln-code"> class="hljs-ln-line"> lm(MD~ T+FC+pH+DOC, mydata),
- class="hljs-ln-numbers"> class="hljs-ln-line hljs-ln-n" data-line-number="3"> class="hljs-ln-code"> class="hljs-ln-line"> lm(MGE~ T+FC+pH+DOC +MD, mydata ),
- class="hljs-ln-numbers"> class="hljs-ln-line hljs-ln-n" data-line-number="4"> class="hljs-ln-code"> class="hljs-ln-line"> lm(ARG~ T+FC+pH+DOC +MD, mydata )
- class="hljs-ln-numbers"> class="hljs-ln-line hljs-ln-n" data-line-number="5"> class="hljs-ln-code"> class="hljs-ln-line">)
- class="hljs-ln-numbers"> class="hljs-ln-line hljs-ln-n" data-line-number="6"> class="hljs-ln-code"> class="hljs-ln-line">summary(model, .progressBar = F)
结果提示模型不合理且缺少路径。

删除不显著的路径并加上提示路径构建模型如下:
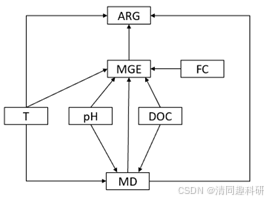
代码如下:
- class="hljs-ln-numbers"> class="hljs-ln-line hljs-ln-n" data-line-number="1"> class="hljs-ln-code"> class="hljs-ln-line">model <- psem(
- class="hljs-ln-numbers"> class="hljs-ln-line hljs-ln-n" data-line-number="2"> class="hljs-ln-code"> class="hljs-ln-line"> lm(MD~ T+pH+DOC, mydata),
- class="hljs-ln-numbers"> class="hljs-ln-line hljs-ln-n" data-line-number="3"> class="hljs-ln-code"> class="hljs-ln-line"> lm(MGE~ T+pH+FC +DOC+MD, mydata ),
- class="hljs-ln-numbers"> class="hljs-ln-line hljs-ln-n" data-line-number="4"> class="hljs-ln-code"> class="hljs-ln-line"> lm(ARG~ T+FC+MD+MGE, mydata )
- class="hljs-ln-numbers"> class="hljs-ln-line hljs-ln-n" data-line-number="5"> class="hljs-ln-code"> class="hljs-ln-line">)
- class="hljs-ln-numbers"> class="hljs-ln-line hljs-ln-n" data-line-number="6"> class="hljs-ln-code"> class="hljs-ln-line">summary(model, .progressBar = F)
#结果合理,最终结果
#AIC值

#模型拟合优度

#路径系数及显著性
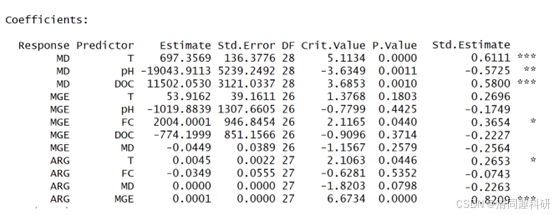
#R2值
#绘图
plot(model,return = FALSE,alpha = 0.05,show = "std") class="hljs-button signin active" data-title="登录复制" data-report-click="{"spm":"1001.2101.3001.4334"}" onclick="hljs.signin(event)">
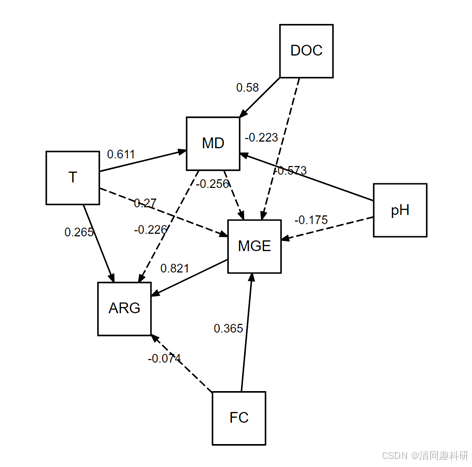
Tips:可通过AI或者PPT进行图片优化。
四、参考文献
Ghosh, S., Matthews, B. & Petchey, O.L. Temperature and biodiversity influence community stability differently in birds and fishes. Nat Ecol Evol 8, 1835–1846 (2024).
Wei, N., Kaczorowski, R.L., Arceo-Gómez, G. et al. Pollinators contribute to the maintenance of flowering plant diversity. Nature 597, 688–692 (2021).
Lefcheck, J.S. (2016), piecewiseSEM: Piecewise structural equation modelling in r for ecology, evolution, and systematics. Methods Ecol Evol, 7: 573-579.
五、相关信息
!!!本文内容由小编总结互联网和文献内容总结整理,如若侵权,联系立即删除!
!!!有需要的小伙伴评论区获取今天的测试代码和实例数据。
📌示例代码中提供了数据和代码,小编已经测试,可直接运行。
以上就是本节全部内容。
如果这篇文章对您有用,请帮忙一键三连(点赞、收藏、评论、分享),让该文章帮助到更多的小伙伴。
data-report-view="{"mod":"1585297308_001","spm":"1001.2101.3001.6548","dest":"https://blog.csdn.net/weixin_46814750/article/details/145394065","extend1":"pc","ab":"new"}">>











评论记录:
回复评论: